- Rigorous screening for researchers and research projects
- Secure shared workspace and file storage
- Airlock: output results securely via the “airlock” and use of SACRO
- Secure Virtual Desktop Infrastructure (VDI)
- GitHub and Gitea
- Linked data “add-ons”
- Sensible pay as you go pricing
- Flexible compute power to suit your needs
- Online “at your own pace” training
- Data specialist support
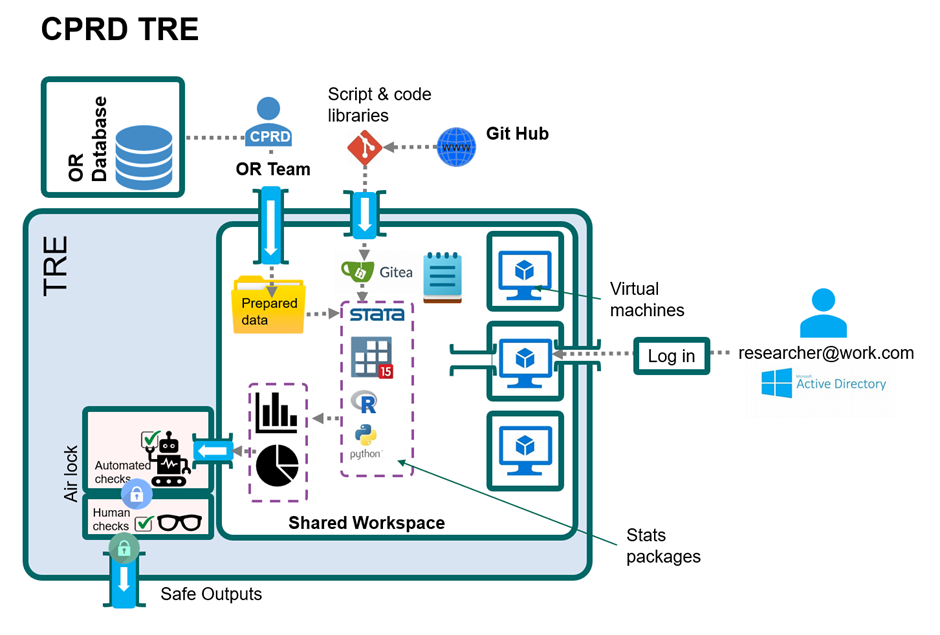
The technical components of the TRE
The CPRD TRE consists of a safe shared workspace that users securely connect to via a virtual machine. The workspace is protected from the internet and provides access to research data, code libraries and analytics tools.
Rigorous screening of researchers and research projects
Before an organisation and its selected researchers can access the CPRD TRE the normal CPRD checks take place:
See Data access.
The research organisation and its researchers sign up to agreements over data use, TRE system use and responsibilities.
Most people find the TRE is intuitive to use. We have also created user guides to support you as you learn how to set up and use the TRE. Checklists are also available as a quick reference.
Secure shared workspace and file storage
Each organisation has its own secure shared workspace where data is stored and statistics packages can be used. The workspace is protected by an “airlock” where checks are made the CPRD research team on data going in or coming out. There are no connections to the internet, although there is a GITea tool available to mirror data from GitHub.
With multiple firewalls and a partition from the internet and main CPRD network, you can trust that the data is safe.
Storage:
Workspace: We provide 5TB of storage within your shared workspace; to cover your primary and linked datasets, applications and analysis outputs. Additional storage can be purchased in packages of 1TB.
Virtual Machine: Each virtual machine has 128GB of storage as part of its operating system (OS) disk.
Airlock: output results securely via the “airlock” and use of SACRO
Outputs have a 3-factor safe data assurance check
- Semi automated checking via the use of SACRO which auto checks outputs for potentially identifying data and visualisations; such as counts less than 10 or scatter plots.
- Human checks by highly qualified epidemiologist researchers.
- Automated file type checks.
Find out more about SACRO: Semi-Automated Checking of Research Outputs.
Secure Virtual Desktop Infrastructure (VDI)
Each researcher is assigned their own Virtual Desktop. It’s like having your own research focused PC inside our safe space to research. Multi-factor authentication protects access. You can work on data within the shared workspace, save files there and collaborate with colleagues; much as you might in your own collaborative environments such as Sharepoint.
Performance:
Tier 1 clients get 4xCPU with 64GB RAM VMs and Tier 2 clients get 4xCPU with 32GB RAM as standard. We’ve based this on testing processing speeds for data sets with populations of 500k and above.
Comprehensive app library inc: Python, RStudio and Stata
As a standard the current Single Study Licence version of the CPRD TRE provides the following statistics software packages: Python, RStudio and Stata. As we develop the TRE towards a full Multi-Study Licence service, we are negotiating with suppliers of the statistics software most commonly used by our clients; to allow broader access to software. Next steps details our roadmap for the future.
Apps installed as standard:
- Anaconda (Python 3.912 64-bit)
- Atlas (Add-on)
- Azul Zulu JRE 8.70.0.23 (8u372) 64-bit
- Azure data studio
- Chrome – to view html - no web access.
- CPRD Code Browser
- Gitea for GitHub
- MS command line utilities 15 for SQL
- Microsoft Visual C++ 2015-2022 (x64 and x86), 2013 (x86)
- MS Visual studio code
- MS Visual Studio tools for applications
- MS ODBC Driver 17 for SQL Server
- MS OLE DB Driver for SQL Server
- MS SQL Server Management Studio 19.1
- Nexus repository
- Notepad++ 64bit x64
- Python Launcher
- R for windows 4.3.0
- R Studio
- R Tools 4.3 5550 5548
- SSMS
- Stata18
MS Notepad is provided within the CPRD TRE as standard, for code and script editing. See also GitHub and Gitea.
GitHub and Gitea
Many researchers use GitHub to store scripts, commands, code libraries and issue tracking for their projects. GitHub is a platform that allows you to create, store, manage, and share your code. It leverages Git software, providing distributed version control along with features like access control, bug tracking, task management, continuous integration, and wikis for any project.
The CPRD TRE uses Gitea to create a safe “pipe in” or mirror of GitHub content, so researchers can access scripts, code, commands and code libraries as flat files; without having to request an import via the airlock. Gitea is one way, so nothing can leave the TRE by this route.
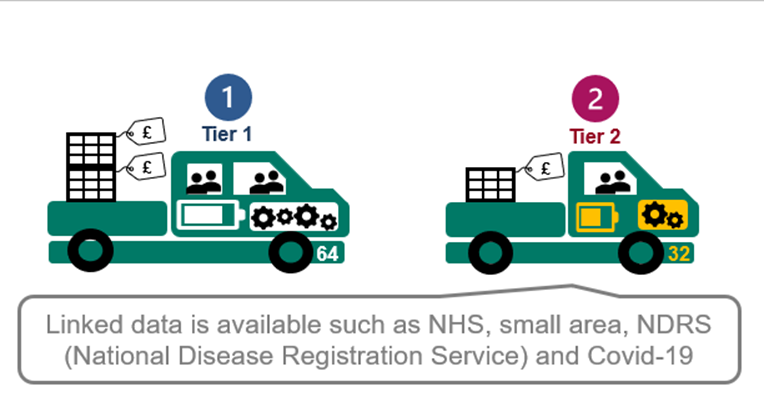
Linked data “add-ons”
If you need additional datasets for your research studies; such as geographic or morbidity specific data, we have many available “add-ons”. These need to be applied for as part of the RDG protocol application process. Post Approval Amendments for “add-ons” are possible in exceptional circumstances.
When a protocol is approved and the dataset required is confirmed, our Observational Research team of epidemiologists will import the data through the airlock to its project or protocol workspace.
As we develop the TRE we will be making core database links available as a standard. For example; our key data sets: ATLAS, CPRD Aurum, and CPRD GOLD.
You can find out more information about these data at Primary care data for public health research and Linked data, and linked data access fees at Pricing.
Sensible pay as you go pricing
This story board explains our pricing model. You can find out more on Pricing under “Single Study Licence”.
1. We offer two tiers of licensing.
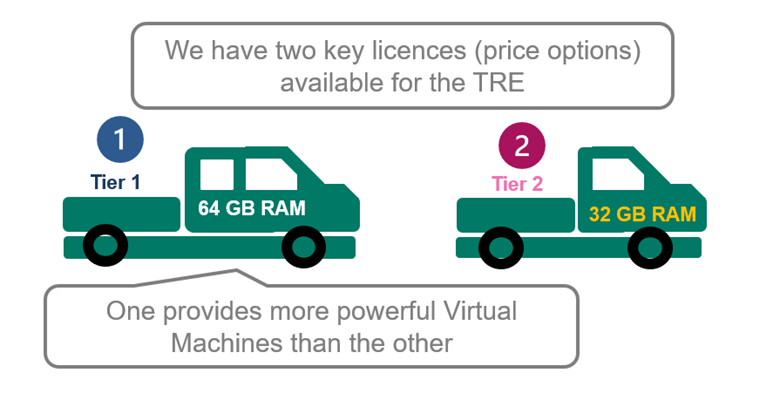
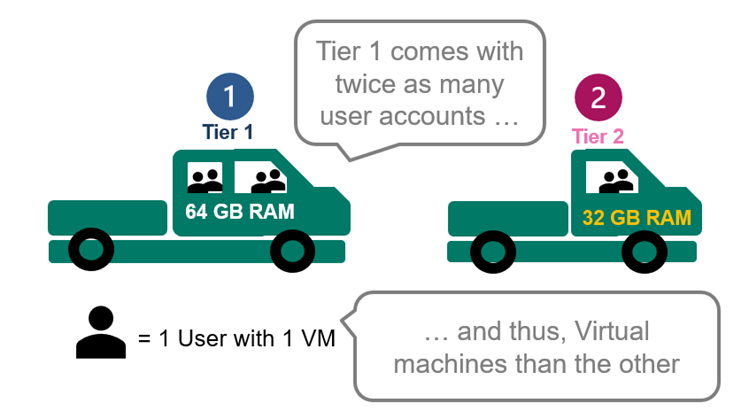
2. Licensing can be purchased according to your needs. Tier 1 comes with 4 user accounts at 64GB RAM and Tier 2 with 2 accounts at 32GB RAM. Both have 4xCPU.
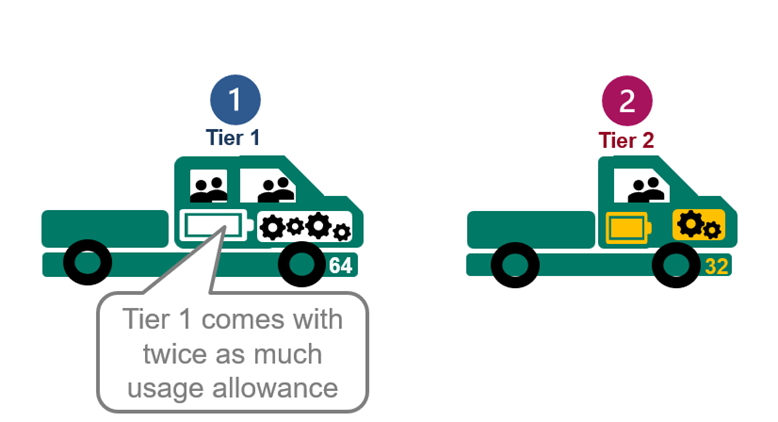
3. Tier 1 users get £4,400 of usage allowance and Tier 2 get £2,200 (exclusive of VAT).
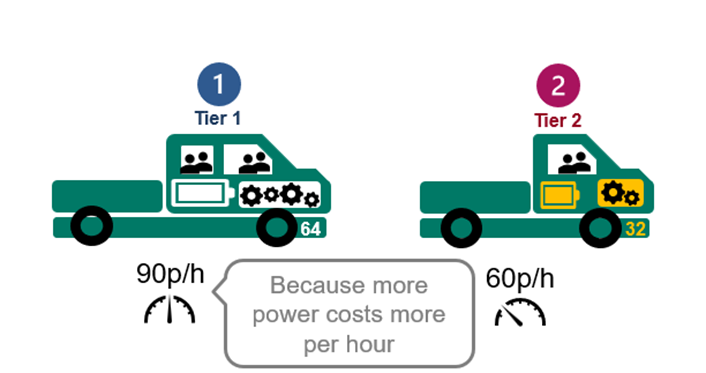
4. Tier 1 VM usage is charged at 90p/hour and Tier 2 at 60p/hr.
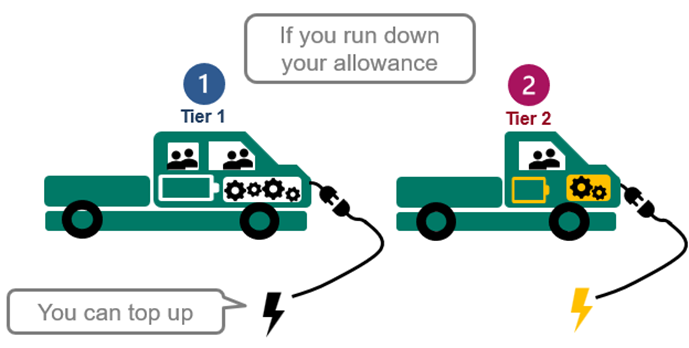
5. You can top up.
6. Pay as you go model.
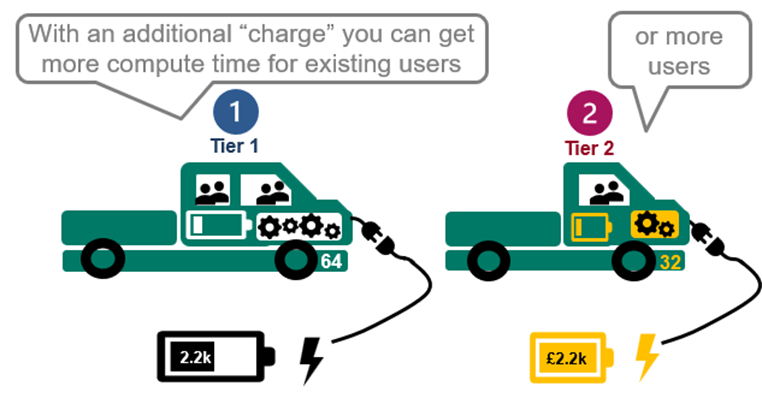
7. Flexible upgrades are possible from your own workspace.
8. You can get “add ons” for specific datasets.
9. Licensing scales, depending on the size of your research and team.
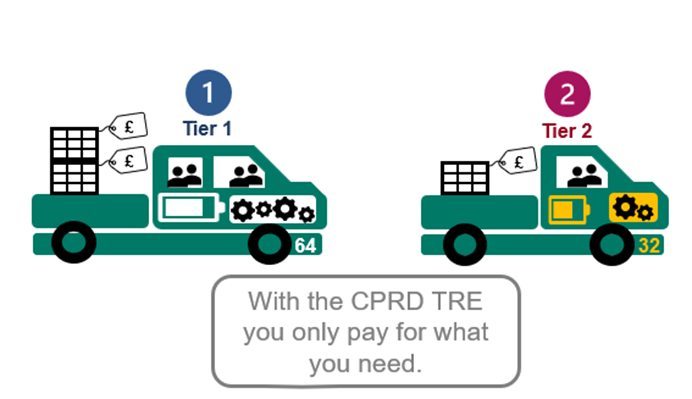
Flexible compute power to suit your needs
With the CPRD TRE you only pay for the computing power that you need. You can upgrade your compute power and number of users. You can also pay for “data add-ons” if you need linked data sets. See the storyboard above.
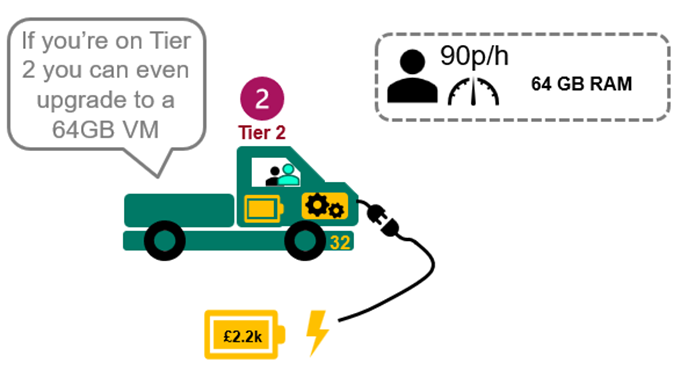
If you have a Tier 2 licence but need a VM on 64GB RAM to ensure your analysis will run, you can upgrade see: Sensible pay as you go pricing.
Research Owners (leads) can upgrade at the click of a button.
Import reference data and code libraries securely
Using the airlock; we can import reference data (code libraries or other categorisation data) that you provide, or data from our selection of linked data “add-ons”. You can also access your code libraries from GitHub.
Online “at your own pace” training
Online in person training will be available to our pilot group. From then on researchers will be able to use our online Training Modules (and later Learning Management System) to learn about how to use the CPRD TRE. Bite size modules and PDF Manuals and will enable researchers to focus on individual learning objectives, building up their understanding at their own pace, whilst providing reference manuals for “on the go” use.
Online training for Python, R-Studio and managing health data is freely available at:
Learn with HDR UK Futures - HDR UK. This site offers an excellent suite of training for data engineering, data science, and data analysis.
Data specialist support
For Tier 1 clients our experienced epidemiologists, statisticians and data scientists support researchers in defining the best data sets for their research projects and protocols as part of the pre-study application. All clients can access support via our contact form within the TRE.
Technical details
OMOP CDM
We are developing the technology to offer access to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) as part of our service OMOP Common Data Model – OHDSI. Our roadmap will keep you up to date on progress.
SQL DB
Whilst initially we will keep our current Single Study Licence model of providing data as a flat file (.csv or text file); we are developing and testing offering Microsoft Open Database Connectivity (ODBC) links to SQL databases (DBs) or providing data cuts as SQL DBs.
Bespoke VM templates
We are consulting with clients on offering further open-source code editing applications beyond Python and R. Approaches to proprietary applications that require a licence are also being investigated.
Population sizes
You use as much storage space and compute power as you need and are charged accordingly. The CPRD TRE is scalable from population sizes from 50k to several million.
CPRD TRE technical support
As the TRE is a secure environment, we have a contact form within the TRE so you can request help and support, whilst providing screen shots of any errors or error messages. Our enquiries team will divert your query to the person who can help the most.